Setup
Overview
Time: minObjectives
Software setup
Please install R and RStudio before this workshop or login to the UIC virtual lab to use the software required for the workshop. See instructions below for both options.
R
R is a programming language that is especially powerful for data exploration, visualization, and statistical analysis. To interact with R, we use RStudio.
Install R by downloading and running this .exe file from CRAN. Also, please install the RStudio IDE. Note that if you have separate user and admin accounts, you should run the installers as administrator (right-click on .exe file and select "Run as administrator" instead of double-clicking). Otherwise problems may occur later, for example when installing R packages.
Video Tutorial
Install R by downloading and running this .pkg file from CRAN. Also, please install the RStudio IDE.
Video Tutorial
Instructions for R installation on various Linux platforms (debian,
fedora, redhat, and ubuntu) can be found at
<https://cran.r-project.org/bin/linux/>. These will instruct you to
use your package manager (e.g. for Fedora run
sudo dnf install R and for Debian/Ubuntu, add a ppa
repository and then run sudo apt-get install r-base).
Also, please install the
RStudio IDE.
Virtual Lab
If you would prefer not to install the software for this workshop on your computer, you may use the Virtual lab service run by Technology Services. This allows you to use a virtual machine either from your web browser or from a desktop app installed on your computer. Overall you may have a better experience using it from the desktop app, but the browswer should suffice for most workshops.
Install the videoconferencing client
If you haven't used Zoom before, go to the official website to download and install the Zoom client for your computer.
Set up your workspace
Like other Carpentries workshops, you will be learning by "coding along" with the Instructors. To do this, you will need to have both the window for the tool you will be learning about (a terminal, RStudio, your web browser, etc..) and the window for the Zoom video conference client open. In order to see both at once, we recommend using one of the following set up options:
- Two monitors: If you have two monitors, plan to have the tool you are learning up on one monitor and the video conferencing software on the other.
- Two devices: If you don't have two monitors, do you have another device (tablet, smartphone) with a medium to large sized screen? If so, try using the smaller device as your video conference connection and your larger device (laptop or desktop) to follow along with the tool you will be learning about.
- Divide your screen: If you only have one device and one screen, practice having two windows (the video conference program and one of the tools you will be using at the workshop) open together. How can you best fit both on your screen? Will it work better for you to toggle between them using a keyboard shortcut? Try it out in advance to decide what will work best for you.
Setup files:
Please download the files emailed to particpate in the workshop:
About the Data Used in this Workshop:
This workshop uses an adapted version of the data paper: Nitsch, F. J., Sellitto, M., & Kalenscher, T. (2021). The effects of acute and chronic stress on choice consistency. Psychoneuroendocrinology, 131, 105289. https://doi.org/10.1016/j.psyneuen.2021.105289.
The data paper along with its underlying data publicly available at: https://osf.io/6mvq7 were adapted and used for educational purposes with authors’ permission.
Key Points
Introduction to R and R studio
Overview
Time: minObjectives
Understand the basics of R and R studio
learn about the Rstudio Interface
R
R is a specialized language most commonly used for statistical computing, data analysis, and implementing graphics. It is open-source and free program. R Language is used worldwide by statisticians and data miners . It helps to perform more efficient and precise data wrangling, analyzing, and visualizing functions for a larger dataset compared to a spreedsheet
Why use R
- R is a very easy and powerful tool for any statistical operations which can also easily be learned by any person even from a non-technical background.
- R offers a variety of packages each of which helps you perform different functions. As of 2021, there are 18839 available packages in R the list of which can be found here.
- R provides a wide variety of statistical (linear and nonlinear modeling, classical statistical tests (e.g., probability, std deviation, etc.), time series analysis, classification, clustering,) and graphical techniques and is highly extensible.
Based on the 2021 survey conducted by Kaggle, R was the third most used programming language by data professionals
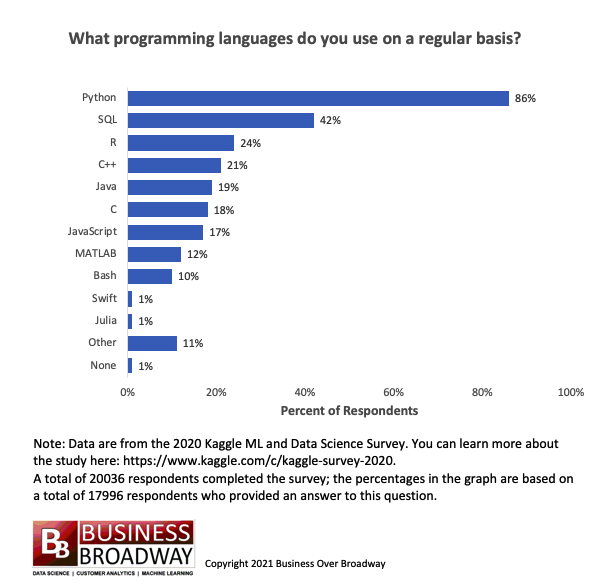
Image Source: Business Broadway, 2021
Understanding R Studio and Console
RStudio is the integrated development environment (IDE) for the basic R software. It is available in two versions:
- RStudio Desktop - Regular desktop application.
- RStudio Server - Runs on a remote server and accessed RStudio using a web browser.
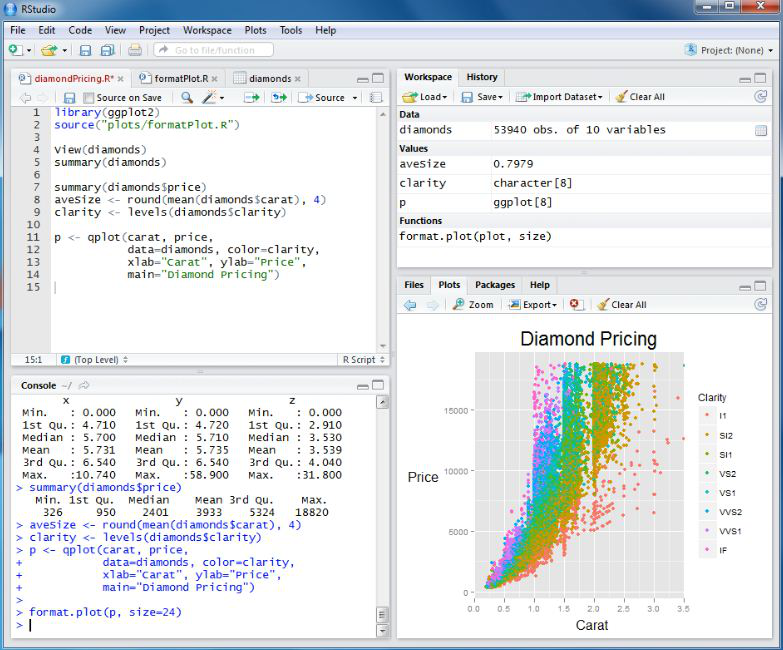
Script Area: - Write codes (or) scripts and run them separately. Also, create a document outline (located on the top right of the script area) in this section that shows all the cod headers in one space.
Console: - Write and run the code together directly here. It also displays the history of any command or an error message in case of a code error.
Environment – List of objects and variables created and present in the current session and also shows the current project file name at the top right of the pane.
Graphics: - Displays the plots, packages, and has an important tab of files. The files option helps us navigate through the different folders of the current project and makes organizing and sorting things a lot better.
The preferences tab in the toolbar helps customize the margins, displays, and font sizes in the r studio.
Help and Cheatsheets in RStudio
help(function_name) – Provides detailed description of function in help window (bottom right) E.g., Run the command help(sort) in the console.
You will now get a complete description of the “sort” function in the help window Points to note:
- If a function’s argument is not given any value (such as x in the above picture) in the help description, this value must be compulsorily specified while running the function
- If a function’s argument is given a value (decreasing = FALSE in above pic) this value is the default value considered by R. It needs to be specified compulsorily when the argument’s value needs to be different.
Cheatsheet – In the wild and woolly world of R there are many packages and to summarize this package functions the cheat sheets come in handy. These cheat sheets are invaluable as learning tools. RStudio has created a large number of cheat sheets, including the one-page R Markdown cheat sheet, which is freely available here
Key Points
R syntax and operators
Overview
Time: 0 minObjectives
perform basic arithematic functions
Understand the basic logical operators
R syntax
Before learning how to code in R, it is necessary to understand the fundamentals of R. As an example, the code below performs basic arithmetic operations such as Addition (+), Subtraction (-), Multiplication (*), Division (/), and Modulo (%%), providing us with output similar to that of a basic calculator. Apart from these simple equations, R is capable of many additional functions for which the following operators are required.
operation directly in the console.
> 2+2
[1] 4
> 2-2
[1] 0
>2*2
[1] 4
> 2/2
[1] 1
> 3%%2
[1] 1
R Console essentials
Loading a Script in R - R provides you with variety of options to run a script. In the Script pane you can either copy and paste a script or type a new script or area select File → Open File → load an existing R Script from the menu.
Run and option command in R - The script’s execution provides us with a code shown in the console pane, which is accessed by clicking the RUN button on the script pane or by pressing the ctrl + Enter keys.On the other hand we can access the options menu by going to Menu bar»tools»options which will provide you with a menu to change the apperance, accessibility and other settings of the R studio.
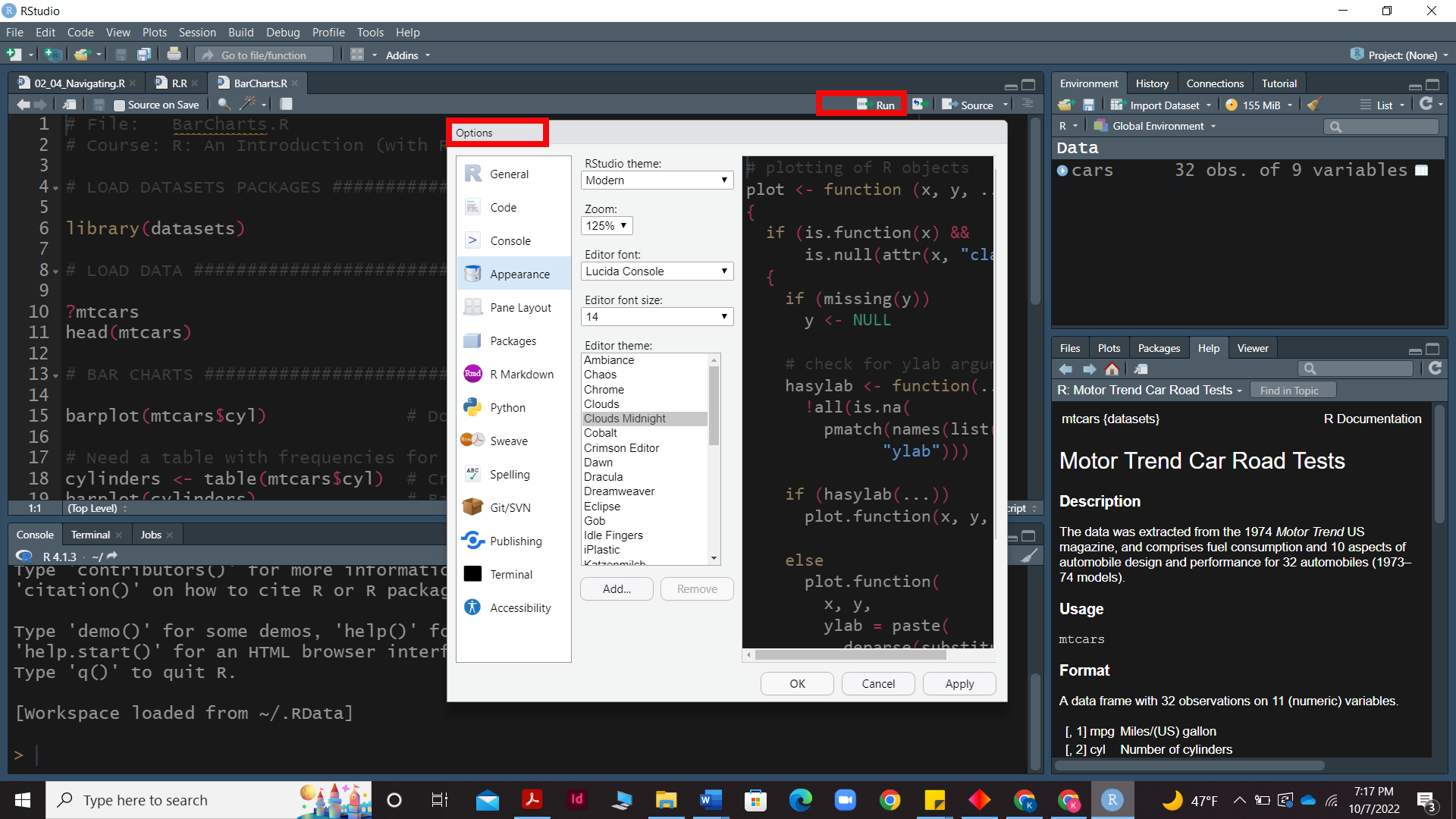
Assigning values in R- Values can be assigned to variables in R using the “<-” symbol. The variable is written on the left and is assigned the value on the right side. For example, to assign a value of 3 to x we can type the code, x <- 3 Assigning values to variables are quite useful especially if these values would be used again. Similar to the previous examples, operations can be performed on the variables to get output directly (or) the output can be stored in a different variable. Once a variable is created it will be visible under the environment section
> x <- 3
> y <- 5
> x+y
[1] 8
> z <- x+y
> z
[1] 8
Commenting in R - Comments are a way to improve your code’s readability and are only meant for the user so the interpreter ignores it. Comments starts with a # When executing the R-code, R will ignore anything that starts with #. Only single-line comments are available in R but we can also use multiline comments by putting then in ‘ or “ quotes. Example:- # This is a comment “Hello World!”
Function in R A key feature of R is functions. Functions are “self contained” modules of code that accomplish a specific task. Functions usually take in some sort of data structure (value, vector, dataframe etc.), process it, and return a result. The general usage for a function is the name of the function followed by parentheses:function_name(input)
One thing to be aware of is that R is case-sensitive. Hence variable “a” is different from “A”
LOGICAL OPERATORS
Provides a list of Boolean results based on operation performed
-
< Less than
-
<= Less than or equal to
-
> Greater than
-
>= Greater than or equal to
-
== Equal to
-
! = Not equal to
-
x&y AND operation
-
x|y OR operation
-
!x NOT operation
Please note that in R the Boolean values “TRUE” & “FALSE” can also be written as “T” &” F”.
Key Points
Variables and datatypes
Overview
Time: minObjectives
understand where to access packages for different functions
learn about the data types permitted for analysis in R studio
Variables
-Variable are basically a storage location where you store some type of value and where that value can be altered based on your need. Variable is also known as Identifier because the variable name identifies the value that is stored in the memory (RAM). As we Know R is a case-sensitive language hence a variable ABC = 15 and Abc= 32 can have different values.
Naming Variables
- Variable name must start with “letter” and can contain a number, letter, underscore (_) and period (‘.’) ```diff
-
variableName1, one.variable ```
- Underscores (_) at the beginning of the variable name is not allowed. ```diff
- _variable_one. ```
- Periods (.) at the beginning of the variable name are allowed but should not be followed by a number eg - ```diff
- .1myvariable ```
-
Reserved words or keywords are not allowed to be identified as a variable name.
- Special characters such as “#”, “&’, etc., along with white spaces (tabs, space) are not allowed in a variable name
Data Types
Data type in R specifies the size and type of information the variable will store.
R language has five main data types
Checking data type in R
There are several functions that can show you the data type of an R object, such as typeof, mode, storage.mode, class and str.the main use of some of them is not to just check the data type of an R object. For instance, the class of an R object can be different from the data type and the str function is designed to show the full structure of an object. If you want to print the R data type, we recommend using the typeof functio There are other functions that allow you to check if some object belongs to some data type, returning TRUE or FALSE. As a rule, these functions start with is. followed by the data type.
Example- is.numeric(4) #true
Data type coercion
You can coerce data types in R with the functions starting with as., summarized
as.numeric == numeric
as.integar == integer
as.double == double
as.character == Character
as.logical== Boolean
as.raw == Raw
Character data type
Character data type stores value or strings and contains alphabets, numbers, and symbols Character data type value is written withing single (‘ ‘)or double inverted quotes (“ “) Example- “A”, “2.21”, “skill@”.
# input code
# Declaring character value with double quotes ""
charac <- "Abcd"
charac
class(charac)
#Convert values to character data type.
pi_value <- 3.14
x <- as.character(pi_value)
x
class(x)
# Concatenation of Character
firstname <- "Kasturi "
lastname <- "Acharya"
# Character Value Concatenation
# Paste function is used to concatenate characters
full_name <- paste (firstname, lastname)
full_name
# output
# Declaring character value with double quotes ""
> charac <- "Abcd"
> charac
[1] "Abcd"
> class(charac)
[1] "character"
> #Convert values to character data type.
> pi_value <- 3.14
> x <- as.character(pi_value)
> x
[1] "3.14"
> class(x)
[1] "character"
>
> # Concatenation of Character
> first_name <- "Kasturi"
> last_name <- "Acharya"
>
> # Character Value Concatenation
> # Paste function is used to concatenate characters
> full_name <- paste (first_name,last_name)
> full_name
[1] "Kasturi Acharya"
Complex data type
- R supports a set of all complex numbers and also stores numbers with an imaginary component. Examples: 1+3i, 5i, 5- 9i
# input code
# Assign complex value to x
x <- 10 + 6i + 20
x
class(x)
z <- 6i
z
class(z)
#Using as.complex() function to convert value to complex.
as.complex(5)
as.complex(7i)
#Performing Addition on Complex Numbers
y1 <- 7+3i
y2 <- 8+9i
sum_y <- y1+y2
sum_y
class(sum_y)
# output
> # Assign complex value to x
> x <- 10 + 6i + 20
> x
[1] 30+6i
> class(x)
[1] "complex"
> z <- 6i
> z
[1] 0+6i
> class(z)
[1] "complex"
> #Using as.complex() function to convert value to complex.
> as.complex(5)
[1] 5+0i
> as.complex(7i)
[1] 0+7i
> #Performing Addition on Complex Numbers
> y1 <- 7+3i
> y2 <- 8+9i
> sum_y <- y1+y2
> sum_y
[1] 15+12i
> class(sum_y)
[1] "complex"
>
Numeric Data Type
- This data type represents all real numbers with or without decimal values, and is most commonly used data type. Example: - 1, 20.5, -97.05, -65
# input code
# Assigning a decimal value to variable x
x <- 15.6
x
class(x)
typeof(x)
x1 <- 20
x1
class(x1)
typeof(x1)
# Converting an integer value to numeric type
x2 <- 22L
class(x2)
typeof(x2)
x3 <- as.numeric(x2)
x3
class(x3)
typeof(x3)
# output
> # Assigning a decimal value to variable x
> x <- 15.6
> x
[1] 15.6
> class(x)
[1] "numeric"
> typeof(x)
[1] "double"
>
> x1 <- 20
> x1
[1] 20
> class(x1)
[1] "numeric"
> typeof(x1)
[1] "double"
>
> # Converting an integer value to numeric type
> x2 <- 22L
> class(x2)
[1] "integer"
> typeof(x2)
[1] "integer"
> x3 <- as.numeric(x2)
> x3
[1] 22
> class(x3)
[1] "numeric"
> typeof(x3)
[1] "double"
Integer Data Type
- Integer data type stores non-decimal values and is denoted with by adding Capital L afer a value.The as. integer () function can be used to convert a number into integer type data in R. Example – 5, 102, 600, 1003.
# input code
x <- 18L # putting capital 'L' after a value forces it to be
# stored as Integer.
class(x)
y <- 9
class(y)
x1 <- 23.0L
x1 <- 23L
class(x1)
# Using integer function to declare an Integer type value
y1 <- as.integer(44)
class(y1)
#coerce a numeric value into integer
y2 <- as.integer(45.2)
y2
#Convert Logical States to Integer
Logic_True <- as.integer(TRUE)
Logic_True
# To check if the value is integer type:
is.integer(x)
is.integer(y)
is.integer(y1)
#Creating integer vector from 1 to 5
m = 1:5
m
class(m)
# output
>
> x <- 18L # putting capital 'L' after a value forces it to be
> # stored as Integer.
> class(x)
[1] "integer"
>
>
> y <- 9
> class(y)
[1] "numeric"
>
>
> x1 <- 23.0L
Warning message:
integer literal 23.0L contains unnecessary decimal point
> x1 <- 23L
> class(x1)
[1] "integer"
>
>
> # Using integer function to declare an Integer type value
> y1 <- as.integer(44)
> class(y1)
[1] "integer"
>
> #coerce a numeric value into integer
> y2 <- as.integer(45.2)
> y2
[1] 45
> #Convert Logical States to Integer
> Logic_True <- as.integer(TRUE)
> Logic_True
[1] 1
> # To check if the value is integer type:
> is.integer(x)
[1] TRUE
> is.integer(y)
[1] FALSE
> is.integer(y1)
[1] TRUE
>
>
> #Creating integer vector from 1 to 5
> m = 1:5
> m
[1] 1 2 3 4 5
> class(m)
[1] "integer"
>
# input code
# BONUS
#Integers value can be a maximum 2147483647 (2 billion)
.Machine$integer.max
#Double value can be a maximum 1.797693e+308 (very much > than 2B)
.Machine$double.xmax
Logical Data Type
- This data type stores logical or Boolean values which are often generated as a result of logical operations. Example – TRUE, FALSE
# input code
x <- TRUE
y<- FALSE
x1 <- T
y1 <- F
typeof(x1)
mode(x1)
####################
# Value Comparison #
####################
# Less Than and Greater Than Comparison
32 < 98 # TRUE Statement
37 > 52 # FALSE Statement
57 == 34 # FALSE Statement
80 == 80 # TRUE Statement
# output
x <- TRUE
> y<- FALSE
>
> x1 <- T
> y1 <- F
> typeof(x1)
[1] "logical"
> mode(x1)
[1] "logical"
> # Value Comparison #
>
> # Less Than and Greater Than Comparison
> 32 < 98 # TRUE Statement
[1] TRUE
> 37 > 52 # FALSE Statement
[1] FALSE
> # Equal TO Comparison
> 57 == 34 # FALSE Statement
[1] FALSE
> 80 == 80 # TRUE Statement
[1] TRUE
Key Points
importance of using packages in R studio for efficient data analysis
Intoduction to strings and data structures
Overview
Time: minObjectives
understand basics of strings and string manipulation
understanding different data structure
learn about functions of each data structure
Strings
- Strings are made of a single character or contain a collection of characters.
- Strings can be created by either single quotes (‘ ‘) or double quotes (“ “)
Rule for String in R
- String starts and ends with a single quote. Double quotes (“ “), and through the escape sequence (‘/’), single quote can become a part of the string. Example- ‘buses’, ‘merry”s’, ‘ merry\’s’
# input code string concatenation
Count number of characters
x1 <- "Olivia"
x2 <- "Jhon"
x3 <- "William"
#checking number of characters
nchar(x1)
nchar(x2)
nchar(x3)
# Letters using vector function in R
# Check the sequence of letters
letters
letters[4]
letters[1:5]
# String Concatenation
# Paste function is used with syntax below:
x <- paste("Hello","World","!",sep = " ")
x
y <- paste(x1,x2,x3,"is happy.")
y
z<- paste("Hello","everyone","!", sep =" ")
z
# Vectors
c() # concatenate function
x4 <- c("Olivia","Jhon","William")
y1 <- paste(x4,"is happy.")
y1
z1 <- c("Please bring me","a few ")
z2 <- c("some vegetables","fruits")
z <- paste(z1,z2,collapse = " and ")
z
# output
# input code string concatenation
> Count number of characters
Error: unexpected symbol in "Count number"
> x1 <- "Olivia"
> x2 <- "Jhon"
> x3 <- "William"
>
> #checking number of characters
> nchar(x1)
[1] 6
> nchar(x2)
[1] 4
> nchar(x3)
[1] 7
>
> # Letters using vector function in R
> # Check the sequence of letters
> letters
[1] "a" "b" "c" "d" "e" "f" "g" "h" "i" "j" "k" "l" "m" "n" "o" "p" "q" "r" "s" "t" "u" "v" "w" "x"
[25] "y" "z"
> letters[4]
[1] "d"
> letters[1:5]
[1] "a" "b" "c" "d" "e"
>
> # String Concatenation
> # Paste function is used with syntax below:
>
> x <- paste("Hello","World","!",sep = " ")
> x
[1] "Hello World !"
>
> y <- paste(x1,x2,x3,"is happy.")
> y
[1] "Olivia Jhon William is happy."
>
> z<- paste("Hello","everyone","!", sep =" ")
> z
[1] "Hello everyone !"
>
> x4 <- c("Olivia","Jhon","William")
> y1 <- paste(x4,"is happy.")
> y1
[1] "Olivia is happy." "Jhon is happy." "William is happy."
>
> z1 <- c("Please bring me","a few ")
> z2 <- c("some vegetables","fruits")
> z <- paste(z1,z2,collapse = " and ")
> z
[1] "Please bring me some vegetables and a few fruits"
String Manipulation
-it’s the process of corecing, slicing, pasting, or analyzing strings
x <- "William is happy today"
x
# Converting all words to upper case using toupper() function
toupper(x)
# Converting all words to lower case using tolower() function
tolower(x)
x1 <- "Henry is A hardworker. He owns A house and A car."
x1
chartr("A", "a", x1)
z <- "I widd gq tq market tqmqrrqw."
chartr("dq","lo", z)
x2 <- "Henry puts in all his good efforts"
x2
substr(x2, start = 22, stop = 27)
#split function
x4 <- "Henry puts in all his good efforts"
class(x4)
y1 <- strsplit(x4, split = " ")
y1
class(y1)
#either create a variable like y1 or direct use the function in case of Mason
strsplit("Mason", split ="")
x4
y2 <- unlist(strsplit(x4, split = " "))
y2
class(y2)
#output
> x <- "William is happy today"
> x
[1] "William is happy today"
>
> # Converting all words to upper case using toupper() function
> toupper(x)
[1] "WILLIAM IS HAPPY TODAY"
>
> # Converting all words to lower case using tolower() function
> tolower(x)
[1] "william is happy today"
>
> x1 <- "Henry is A hardworker. He owns A house and A car."
> x1
[1] "Henry is A hardworker. He owns A house and A car."
> chartr("A", "a", x1)
[1] "Henry is a hardworker. He owns a house and a car."
>
> z <- "I widd gq tq market tqmqrrqw."
> chartr("dq","lo", z)
[1] "I will go to market tomorrow."
>
> x2 <- "Henry puts in all his good efforts"
> x2
[1] "Henry puts in all his good efforts"
> substr(x2, start = 22, stop = 27)
[1] " good "
>
> #split function
>
> x4 <- "Henry puts in all his good efforts"
> class(x4)
[1] "character"
> y1 <- strsplit(x4, split = " ")
> y1
[[1]]
[1] "Henry" "puts" "in" "all" "his" "good" "efforts"
> class(y1)
[1] "list"
> #either create a variable like y1 or direct use the function in case of Mason
> strsplit("Mason", split ="")
[[1]]
[1] "M" "a" "s" "o" "n"
> x4
[1] "Henry puts in all his good efforts"
> y2 <- unlist(strsplit(x4, split = " "))
> y2
[1] "Henry" "puts" "in" "all" "his" "good" "efforts"
> class(y2)
[1] "character"
Data Structure
A data structure is essentially a way to organize data in a system to facilitate effective usage of the same.Data structures are the objects that are manipulated regularly in R. They are used to store data in an organized fashion to make data manipulation and other data operations more efficient. R has many data structure which are as follows
- Vectors
- Matrices
- Factors
- Data Frames
- Arrays
- Lists
Vector
Vectors are the basic data structure of R. Vectors can hold multiple values together using the concatenate c() function. The type of data inside a vector can be determined by using the type of() function and the length (or) number of elements in a vector can be found with the length() function. R uses one indexing unlike python, hence the position of the first component in a vector can be accessed by vector name [1] A vector will always contain data of the same data type. If a vector contains multiple data types the vector will convert all its values to the same data type in the below order of precedence:
- Character
- Double (Float / Decimals)
- Integers (Round whole numbers)
Technically, vectors can be one of two types:
- atomic vectors
- lists although the term “vector” most commonly refers to the atomic types not to lists.
Matrices and Array
Adding a dim attribute to an atomic vector allows it to behave like a multi-dimensional array. A special case of the array is the matrix, which has two dimensions. Matrices are used commonly as part of the mathematical machinery of statistics. Arrays are much rarer, but worth being aware of. Matrices and arrays are created with matrix() and array(), or by using the assignment form of dim()
Data Frames
A data frame is a very important data type in R. It’s pretty much the de facto data structure for most tabular data and what we use for statistics (explained in detail in the next section) Some additional information on data frames: Usually created by read.csv() and read.table(), i.e. when importing the data into R. Assuming all columns in a data frame are of same type, data frame can be converted to a matrix with data.matrix() (preferred) or as.matrix(). Otherwise type coercion will be enforced and the results may not always be what you expect.
You can also create a new data frame with data.frame() function. Find the number of rows and columns with nrow(dat) and ncol(dat), respectively. Rownames are often automatically generated and look like 1, 2, …, n. Consistency in numbering of rownames may not be honored when rows are reshuffled or subset.
Lists
In R lists act as containers. Unlike atomic vectors, the contents of a list are not restricted to a single mode and can encompass any mixture of data types. Lists are sometimes called generic vectors, because the elements of a list can by of any type of R object, even lists containing further list. You can create lists using list() or coerce other objects using as.list()
# DATA STRUCTURES ##########################################
## Vector ##################################################
v1 <- c(1, 2, 3, 4, 5)
v1
is.vector(v1)
v2 <- c("a", "b", "c")
v2
is.vector(v2)
v3 <- c(TRUE, TRUE, FALSE, FALSE, TRUE)
v3
is.vector(v3)
## Matrix ##################################################
m1 <- matrix(c(T, T, F, F, T, F), nrow = 2)
m1
m2 <- matrix(c("a", "b",
"c", "d"),
nrow = 2,
byrow = T)
m2
## Array ###################################################
# Give data, then dimemensions (rows, columns, tables)
a1 <- array(c( 1:24), c(4, 3, 2))
a1
## Data frame ##############################################
# Can combine vectors of the same length
vNumeric <- c(1, 2, 3)
vCharacter <- c("a", "b", "c")
vLogical <- c(T, F, T)
dfa <- cbind(vNumeric, vCharacter, vLogical)
dfa # Matrix of one data type
df <- as.data.frame(cbind(vNumeric, vCharacter, vLogical))
df # Makes a data frame with three different data types
## List ####################################################
o1 <- c(1, 2, 3)
o2 <- c("a", "b", "c", "d")
o3 <- c(T, F, T, T, F)
list1 <- list(o1, o2, o3)
list1
list2 <- list(o1, o2, o3, list1) # Lists within lists!
list2
# COERCING TYPES ###########################################
## Automatic coercion ######################################
# Goes to "least restrictive" data type
(coerce1 <- c(1, "b", TRUE))
# coerce1 # Parenthese around command above make this moot
typeof(coerce1)
## Coerce numeric to integer ###############################
(coerce2 <- 5)
typeof(coerce2)
(coerce3 <- as.integer(5))
typeof(coerce3)
## Coerce character to numeric #############################
(coerce4 <- c("1", "2", "3"))
typeof(coerce4)
(coerce5 <- as.numeric(c("1", "2", "3")))
typeof(coerce5)
## Coerce matrix to data frame #############################
(coerce6 <- matrix(1:9, nrow= 3))
is.matrix(coerce6)
(coerce7 <- as.data.frame(matrix(1:9, nrow= 3)))
is.data.frame(coerce7)
# CLEAN UP #################################################
# Clear environment
rm(list = ls())
# Clear console
cat("\014") # ctrl+L
# Clear mind :)
Key Points
understanding strings and vectors
Introduction to data frame
Overview
Time: 0 minObjectives
learn how to create and access a data frame
learn data frame transformation and operations
Data Frames
Data frames are used for storing Data tables in R. They are two-dimensional array structures and are similar to tables where each column represents one variable. The main features to note about a data frame are:
- Columns can be of different data types
- Each column name must be unique
- Each column should be of the same length i.e., contain the same number of elements
Data frames in R can be created in two ways:
- Using data.frame() command
- Importing data from files such as .csv, .xlsx etc.
data.frame() FUNCTION:
While using the command we can follow the below syntax
data. Frame (column_1, column_2, column_3, …………………….)
Make sure that the names of the columns are unique and are of the same length.
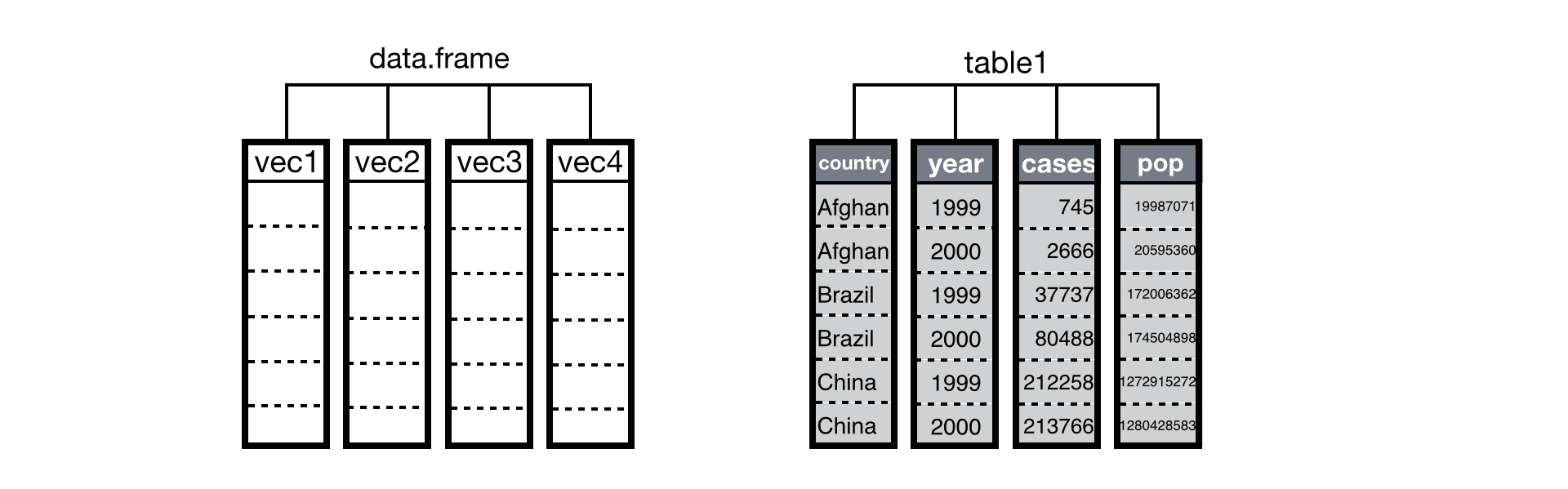
Creating a data frame
# input code
# Student ID, names and their marks.
student.data <- data.frame(
std_id = c(001:005),
std_name = c("William", "James", "Olivia", "Steve", "David"),
std_marks = c(84.8, 98.4, 74.6, 80, 95)
)
# Display the dataframe student.data
student.data
# Check the structure of the dataframe student.data
str(student.data)
#check the head and tail of the dataframe student.data
head(student.data, 3)
tail(student.data, 3)
# Check the summary, lenth and dimension of the dataframe student.data
summary(student.data)
length(student.data)
dim(student.data)
# Check number of row/columns individually.
ncol(student.data)
nrow(student.data)
#output
> # Student ID, names and their marks.
> student.data <- data.frame(
+ std_id = c(001:005),
+ std_name = c("William", "James", "Olivia", "Steve", "David"),
+ std_marks = c(84.8, 98.4, 74.6, 80, 95)
+ )
>
> # Display the dataframe student.data
> student.data
std_id std_name std_marks
1 1 William 84.8
2 2 James 98.4
3 3 Olivia 74.6
4 4 Steve 80.0
5 5 David 95.0
>
> # Check the structure of the dataframe student.data
> str(student.data)
'data.frame': 5 obs. of 3 variables:
$ std_id : int 1 2 3 4 5
$ std_name : chr "William" "James" "Olivia" "Steve" ...
$ std_marks: num 84.8 98.4 74.6 80 95
>
> #check the head and tail of the dataframe student.data
> head(student.data, 3)
std_id std_name std_marks
1 1 William 84.8
2 2 James 98.4
3 3 Olivia 74.6
>
> tail(student.data, 3)
std_id std_name std_marks
3 3 Olivia 74.6
4 4 Steve 80.0
5 5 David 95.0
>
>
> # Check the summary, lenth and dimension of the dataframe student.data
> summary(student.data)
std_id std_name std_marks
Min. :1 Length:5 Min. :74.60
1st Qu.:2 Class :character 1st Qu.:80.00
Median :3 Mode :character Median :84.80
Mean :3 Mean :86.56
3rd Qu.:4 3rd Qu.:95.00
Max. :5 Max. :98.40
>
> length(student.data)
[1] 3
>
> dim(student.data)
[1] 5 3
>
> # Check number of row/columns individually.
> ncol(student.data)
[1] 3
> nrow(student.data)
[1] 5
Importing data
There are multiple commands with various arguments to import data from different file formats into R environment. I shall show the simplest command to import a csv file as a data frame
data_frame_name <- read.csv(file. choose(), header = T) Here, file. choose() - Allows you to choose a .csv file stored in your local desktop Here, header = T - Indicates the first row in the file contains column names.
Double click (or) click once and select open on your desired file to import Once the data has been imported successfully the data frame would be visible with its name in the Environment pane on the top right.
Packages
- One of the most important things in R is its collection of Packages. The package is a collection of R functions, data, and compiled code and Library is the location where the packages are stored. In order to access these packages, we can either go to r-project. Org > CRAN> 0 Cloud> packages>CRAN task view or use the command library() to load the package in the current R session.
- Then just call the appropriate package functions
install.packages(“package_name”) – Install the package from CRAN repository
install.packages( c(“package_1”, “”package_2”, “package_3”) ) -Install multiple packages
library(“package_name”) – Load the package in current R session.
Importing dataset and Packages
getwd()
Install Package############################
# I recommend "pacman" for managing add-on packages. It will
# install packages, if needed, and then load the packages.
install.packages("pacman")
# Then load the package by using either of the following:
require(pacman) # Gives a confirmation message.
library(pacman) # No message.
# Or, by using "pacman::p_load" you can use the p_load
# function from pacman without actually loading pacman.
# These are packages I load every time.
pacman::p_load(pacman, dplyr, GGally, ggplot2, ggthemes,
ggvis, httr, lubridate, plotly, rio, rmarkdown, shiny,
stringr, tidyr)
library(datasets) # Load/unload base packages manually
Using Tidyverse#############################################
install.packages("tidyverse")
library (tidyverse)
Importing Data #############################################
df <- read.csv("StateData.csv")
df
head(df)
str(df)
summary (df)
df[c("State", "governor")]
head(df[c("State", "governor")])
summary(df[c("State", "governor")])
sum (df[c("State", "governor")]))
df = df.sum(axis=1)
df1 <- c(sum(df$instagram), sum(df$facebook))
df1
c (sum(df$instagram), sum(df$retweet))
sum(df$State) # character datatype
mean(df$instagram)
"1" %in% df$instagram
Key Points
basic statistical knowledge and formulas
sample dataset and importing data in R studio
Overview
Time: minObjectives
learning how to use the sample dataset
understanding how to import data in R studio
Sample Dataset
- One of the easiest ways to start experimenting with the analysis in R is by means of built-in sample datasets available in R. These datasets are available in their own package.
- Use the code provided in the script to load the dataset and then toggle through help function to know the complete information of the dataset.
# INSTALL AND LOAD PACKAGES ################################
# Load base packages manually
library(datasets) # For example datasets
?datasets
library(help = "datasets")
# SOME SAMPLE DATASETS #####################################
iris
?iris
cars <-cars
head(cars)
iris <- iris
head(iris)
tail(iris,20)
iris[,c(1,2)]
iris[,c('Sepal.Length')]
str(iris)
rm(list = ls())
iris
# CLEAN UP #################################################
# Clear environment
rm(list = ls())
# Clear packages
detach("package:datasets", unload = TRUE) # For base
# Clear plots
dev.off() # But only if there IS a plot
# Clear console
cat("\014") # ctrl+L
Data Visulaistion using the basic Plot function in R
The most used plotting function in R programming is the plot() function. It is a generic function, meaning, it has many methods which are called according to the type of object passed to plot(). In the simplest case, we can pass in a vector and we will get a scatter plot (default) of magnitude vs index. But generally, we pass in two vectors and a scatter plot of these points are plotted. For example, the command plot(c(1,2),c(3,5)) would plot the points (1,3) and (2,5) In the exercise below we will see how to create a generic plot, bar chart and a histogram in R using the Sample dataset available in the Program
PLOT FUNCTION IN R
LOAD DATASETS PACKAGES ###################################
library(datasets) # Load/unload base packages manually
LOAD DATA ################################################
head(iris)
PLOT DATA WITH PLOT() ####################################
?plot # Help for plot()
plot(iris$Species) # Categorical variable
plot(iris$Petal.Length) # Quantitative variable
plot(iris$Species, iris$Petal.Width) # Cat x quant
plot(iris$Petal.Length, iris$Petal.Width) # Quant pair
plot(iris) # Entire data frame
Plot with options
plot(iris$Petal.Length, iris$Petal.Width,
col = "#cc0000", # Hex code for datalab.cc red
pch = 19, # Use solid circles for points
main = "Iris: Petal Length vs. Petal Width",
xlab = "Petal Length",
ylab = "Petal Width")
PLOT FORMULAS WITH PLOT() ################################
plot(cos, 0, 2*pi)
plot(exp, 1, 5)
plot(dnorm, -3, +3)
Formula plot with options
plot(dnorm, -3, +3,
col = "#cc0000",
lwd = 5,
main = "Standard Normal Distribution",
xlab = "z-scores",
ylab = "Density")
Plotting Bar Chart in R
library(datasets)
LOAD DATA ###############################################
?mtcars
head(mtcars)
sort.default(mtcars, decreasing = TRUE)
BAR CHARTS ###############################################
barplot(mtcars$cyl) # Doesn't work
Need a table with frequencies for each category
cylinders <- table(mtcars$cyl) # Create table
barplot(cylinders) # Bar chart
plot(cylinders) # Default X-Y plot (lines)
CLEAN UP #################################################
Clear environment
rm(list = ls())
Clear packages
detach("package:datasets", unload = TRUE) # For base
Clear plots
dev.off() # But only if there IS a plot
Clear console
cat("\014") # ctrl+L
Plotting Historgram in R
LOAD PACKAGES ############################################
library(datasets)
LOAD DATA ################################################
?iris
head(iris)
BASIC HISTOGRAMS #########################################
hist(iris$Sepal.Length)
hist(iris$Sepal.Width)
hist(iris$Petal.Length)
hist(iris$Petal.Width)
HISTOGRAM BY GROUP #######################################
Put graphs in 3 rows and 1 column
par(mfrow = c(3, 1))
Histograms for each species using options
hist(iris$Petal.Width [iris$Species == "setosa"],
xlim = c(0, 3),
breaks = 9,
main = "Petal Width for Setosa",
xlab = "",
col = "red")
hist(iris$Petal.Width [iris$Species == "versicolor"],
xlim = c(0, 3),
breaks = 9,
main = "Petal Width for Versicolor",
xlab = "",
col = "purple")
hist(iris$Petal.Width [iris$Species == "virginica"],
xlim = c(0, 3),
breaks = 9,
main = "Petal Width for Virginica",
xlab = "",
col = "blue")
Restore graphic parameter
par(mfrow=c(1, 1))
CLEAN UP #################################################
Clear packages
detach("package:datasets", unload = TRUE) # For base
Clear plots
dev.off() # But only if there IS a plot
Clear console
cat("\014") # ctrl+L
Clear mind :)
This brings us to an end of this workshop however the reference section provides links to all the materials used in this workshop and links which provide more detailed understanding of Each Package in R.
Key Points